What’s up with all this Bayesian statistics bs?
This post is an adaptation of a talk I gave at the Columbus Georgia Data Science Meetup .
Example 1: The BS Disease
Uh oh! You just learned that you tested positive for the very serious BS disease . It affects 0.1% of the population. The test for the disease is 99% accurate. What are the chances that you actually have BS?
Bayes’ Theorem
\[ P(BS \mid test) = \frac{P(test \mid BS) \, P(BS)}{P(test)} \]
The test is 99% accurate, so \(P(test \mid BS) = 0.99\) . BS prevalence is 0.1%, so \(P(BS) = 0.001\) . We want to know chance of testing positive, \(P(test)\) , so we reformulate like so:
\[P(test) = P(BS) \times P(test \mid BS) + P(-BS) \times P(test \mid -BS)\]
So we plug and chug:
\[P(BS \mid test) = \frac{0.99 \times 0.001}{0.001 \times 0.99 + (1 - 0.001) \times (1 - 0.99)}\]
You tested positive, but there’s only a 9% chance that you actually have BS.
What’s the point?
Bayes’ Theorem rocks!
\[
\begin{aligned}
\overbrace{p(\theta | \text{Data})}^{\text{posterior}} &= \frac{\overbrace{p(\text{Data} | \theta)}^{\text{likelihood}} \times \overbrace{p(\theta)}^{\text{prior}}}{\underbrace{p(\text{Data})}_{\text{marginal likelihood}}} \\
&\propto p(\text{Data} | \theta) \times p(\theta) \\
\end{aligned}
\]
We can confront our model with the data and incorporate our prior understanding to draw inference about parameters of interest.
Example 2: A/B Testing
Stan
Probabilistic programming language
Implements fancy HMC algorithms
Written in C++ for speed
Interfaces for R (rstan), python (pystan), etc.
Active developers on cutting-edge of HMC research
Large user community
A/B Testing
Suppose your manager approaches you and says,
We’re testing an exciting new web feature! Version A has a red button, and version B has a green button. Which version yields more conversions?”
How can you answer this question?
The Data
<- expand.grid (version = c ("A" , "B" ), converted = c ("Yes" , "No" )$ n_visitors <- c (1300 , 1275 , 120 , 125 )
version converted n_visitors
1 A Yes 1300
2 B Yes 1275
3 A No 120
4 B No 125
The Model
The model of the experiment is an A/B test in which
\[
\begin{aligned}
n_A &\sim \mathsf{Binomial}(N_A, \pi_A) & n_B &\sim \mathsf{Binomial}(N_B, \pi_B) \\
\pi_A &= \mathsf{InvLogit}(\eta_A) & \pi_B &= \mathsf{InvLogit}(\eta_B) \\
\eta_A &= \mathsf{Normal}(0, 2.5) & \eta_B &= \mathsf{Normal}(0, 2.5)
\end{aligned}
\]
Implementation in Stan
data {int <lower =1 > visitors_A;int <lower =0 > conversions_A;int <lower =1 > visitors_B;int <lower =0 > conversions_B;parameters {real eta_A;real eta_B;transformed parameters {real <lower =0. , upper =1. > pi_A = inv_logit(eta_A);real <lower =0. , upper =1. > pi_B = inv_logit(eta_B);model {0. , 2.5 );0. , 2.5 );generated quantities {real <lower =-1. , upper =1. > pi_diff;real eta_diff;real lift;
Run and fit the model
<- list (visitors_A = 1300 ,visitors_B = 1275 ,conversions_A = 120 ,conversions_B = 125 <- rstan:: sampling (data = abtest_data,chains = 1 , iter = 1000 , refresh = 0
Calculate the probability that \(\pi_A\)
<- drop (rstan:: extract (abtest_fit, "pi_A" )[[1 ]])<- drop (rstan:: extract (abtest_fit, "pi_B" )[[1 ]])mean (pi_A > pi_B)
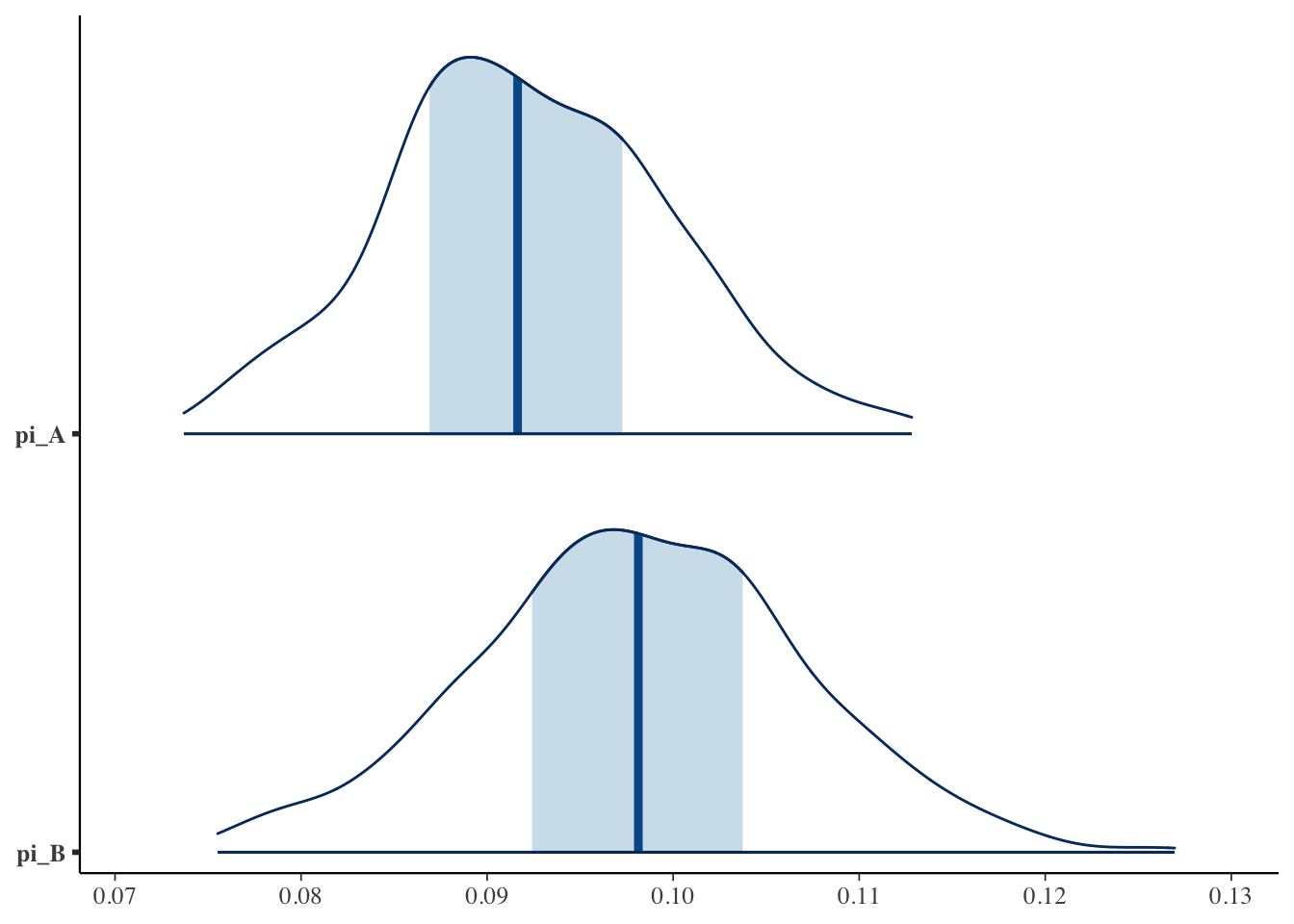
Looks like Version B wins!
Plot posteriors
<- as.matrix (abtest_fit):: mcmc_areas (posterior, pars = c ("pi_A" , "pi_B" ))